- My personal website/
- Teaching Portfolio/
- Corporate strategy in a network perspective/
- Session 1: Introduction to network analysis/
Session 1: Introduction to network analysis

Table of Contents
Session 1 - Introduction to network analysis
The first thing you need to do is to set a project folder called virksomhedsstrategi which you can either place on your desktop, or into a another folder (such as one called 6_semester).
In this folder, I would like you to create 4 new sub-folders. Make sure that all of these are spelled with small caps, to make typing easier.
-
a folder called
inputwhere you will all the data files (incsv,xlsx, or other formats) -
a folder called
outputwhere you will save all network figures we will produce -
a folder called
rwhere you will save all your r-scripts. Make sure to make an r-script for each of the sessions.
After you have created all of the folders, you can open your Rstudio. Create a new r-script, and set your working directory to the file virksomhedsstrategi.
For macOs users, click on the folder name, and press Option + CMD + C. Paste the path into the function setwd(). It should look something similar to this:
setwd("/Users/alexandergamerdinger/Library/CloudStorage/OneDrive-CBS-CopenhagenBusinessSchool/PhD/teaching/virksomhedsstrategi_2023")
1.1 Installing and loading packages
After you have set your working directory, we will install and load important packages that we will use throughout the course.
-
The
tidyversepackage, which is basically a collection of a variety of packages that makes it easy and visually pleasing to manipulate and work with data -
The
igraphpackage, which allows us to construct network objects -
The
ggraphpackage, which enables us to create beautiful network graphs -
The
data.tablepackage, which we will use occasionally for some data wrangling
How do we install and load packages? Please note that packages need to only be installed once. This means that you can simply delete the install.packages() function right after its execution.
# install packages (but only run this once)
install.packages("tidyverse")
install.packages("igraph")
install.packages("ggraph")
install.packages("data.table")
# load pckages (run this every time you open/re-open your r-script)
library("tidyverse")
library("igraph")
library("ggraph")
library("data.table")
1.1.1 A note on the tidyverse syntax
The tidyverse package is great because it has transformed the way of writing R, and has made it much more beautiful. One very important aspect of the package is the use of the pipe %>%.
The pipe allows us to write cleaner and more visually pleasing code. Let us take a look at this data set iris with the attempt to find the mean petal length and width of all species above a petal width of 0.2.
In baseR you would have to write this:
#finding a sub set
subset <- subset(iris, Petal.Width > 0.2)
# selecting only columns on Petals
only_pedal <- subset[,c("Petal.Length", "Petal.Width", "Species")]
# finding mean Petal length and width per species
aggregate(subset[ ,c("Petal.Length", "Petal.Width")],
by = list(subset$Species),
FUN = mean)
## Group.1 Petal.Length Petal.Width
## 1 setosa 1.51875 0.375
## 2 versicolor 4.26000 1.326
## 3 virginica 5.55200 2.026
In dplyr which is a sub-package of tidyverse, you would only have to write this:
iris %>%
# finding a sub set
filter(Petal.Width > 0.2) %>%
# selecting only three columns
select(Petal.Length, Petal.Width, Species) %>%
#grouping by species
group_by(Species) %>%
# aggregating by mean
summarize(across(starts_with("Petal"), mean))
## # A tibble: 3 × 3
## Species Petal.Length Petal.Width
## <fct> <dbl> <dbl>
## 1 setosa 1.52 0.375
## 2 versicolor 4.26 1.33
## 3 virginica 5.55 2.03
The pipe operator allows you to simply pass the output of one function as an input to another function that comes afterwards. This makes your code easily readable and efficient.
If you want to know more about the pipe operator, see this website here: https://uc-r.github.io/pipe
1.2 Loading a data set
After you have set your working directory, and loaded all the required packages, we can now load the data set called den17-no-nordic-letters.csv. This data set was made by Christoph and Anton and can also be found on Github.
First, download the data set and place it into the virksomhedsstrategi/input folder.
Second, load the .csv data file with the function read_csv(). Remember to load your data set from your current working directory.
# loading data | If there is an error,
#it is probably because of your working directory
den <- read_csv("input/den17-no-nordic-letters.csv")
## Rows: 56849 Columns: 17
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (8): name, affiliation, role, tags, sector, type, description, gender
## dbl (6): position_id, id, cvr_person, cvr_affiliation, person_id, affiliati...
## dttm (3): created, archived, last_checked
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# look at the head of the data set
head(den)
## # A tibble: 6 × 17
## name affiliation role tags position_id id sector type description
## <chr> <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
## 1 Aage Almt… Middelfart… Memb… Corp… 1 95023 Corpo… <NA> Automatisk…
## 2 Aage B. A… Foreningen… Memb… Char… 4 67511 NGO Orga… Direktoer
## 3 Aage Chri… AARHUS SOE… Chai… Foun… 6 100903 Found… <NA> Automatisk…
## 4 Aage Dam Branchefor… Chai… Busi… 8 69156 NGO Orga… Formand, A…
## 5 Aage Dam Dansk Erhv… Memb… Empl… 9 72204 NGO Stat Adm. dir. …
## 6 Aage Fran… Dommere va… Memb… Judg… 15 73158 Parli… <NA> <NA>
## # ℹ 8 more variables: created <dttm>, archived <dttm>, last_checked <dttm>,
## # cvr_person <dbl>, cvr_affiliation <dbl>, person_id <dbl>,
## # affiliation_id <dbl>, gender <chr>
1.3 Data manipulation with dplyr
1.3.1 Summary statistics
The data set den has 56,849 rows of individuals and 17 columns.
dim(den) # we could also write: den %>% dim()
## [1] 56849 17
We use the following functions in order to gain a better overview of our data set:
-
the
glimpse()function to understand the overall structure of the data set -
the
count()function to get summary statistics of one or several variables -
the
view()function to see the whole data set, or a subset, on the big screen.
glimpse(den) # we could also write: den %>% glimpse()
## Rows: 56,849
## Columns: 17
## $ name <chr> "Aage Almtoft", "Aage B. Andersen", "Aage Christensen"…
## $ affiliation <chr> "Middelfart Sparekasse", "Foreningen OEstifterne - Rep…
## $ role <chr> "Member", "Member", "Chairman", "Chairman", "Member", …
## $ tags <chr> "Corporation, FINA, Banks, Finance", "Charity, Foundat…
## $ position_id <dbl> 1, 4, 6, 8, 9, 15, 28, 30, 32, 34, 38, 41, 47, 49, 58,…
## $ id <dbl> 95023, 67511, 100903, 69156, 72204, 73158, 100249, 316…
## $ sector <chr> "Corporations", "NGO", "Foundations", "NGO", "NGO", "P…
## $ type <chr> NA, "Organisation", NA, "Organisation", "Stat", NA, NA…
## $ description <chr> "Automatisk CVR import at 2016-03-12 18:01:28: BESTYRE…
## $ created <dttm> 2016-03-12 18:01:28, 2016-02-05 14:45:10, 2016-03-12 …
## $ archived <dttm> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ last_checked <dttm> 2017-11-09 15:38:01, 2016-02-12 14:41:09, 2017-11-09 …
## $ cvr_person <dbl> 4003983591, NA, 4000054465, NA, NA, NA, 4003907021, NA…
## $ cvr_affiliation <dbl> 24744817, NA, 29094411, NA, 43232010, NA, 25952200, NA…
## $ person_id <dbl> 1, 3, 4, 5, 5, 9, 16, 18, 20, 21, 23, 25, 30, 31, 36, …
## $ affiliation_id <dbl> 3687, 2528, 237, 469, 1041, 1781, 4878, 1038, 3535, 27…
## $ gender <chr> "Men", "Men", "Men", "Men", "Men", "Men", "Men", "Men"…
If we are interested in seeing how many data entries there are per sector, we can write the following command.
den %>% # here, we start to use the pipe
count(sector) # this gives us an unordered summary statistics
## # A tibble: 13 × 2
## sector n
## <chr> <int>
## 1 Commissions 795
## 2 Corporations 7989
## 3 Events 1948
## 4 Family 207
## 5 Foundations 6987
## 6 Municipal 320
## 7 NGO 17720
## 8 Organisation 6
## 9 Parliament 1087
## 10 Politics 37
## 11 State 13601
## 12 VL_networks 3803
## 13 <NA> 2349
den %>%
count(sector, sort = TRUE) # this gives us the same thing, but ordered
## # A tibble: 13 × 2
## sector n
## <chr> <int>
## 1 NGO 17720
## 2 State 13601
## 3 Corporations 7989
## 4 Foundations 6987
## 5 VL_networks 3803
## 6 <NA> 2349
## 7 Events 1948
## 8 Parliament 1087
## 9 Commissions 795
## 10 Municipal 320
## 11 Family 207
## 12 Politics 37
## 13 Organisation 6
1.3.1 Data subsets
Most of the time, we are interested in looking at subsets of the data set den, e.g. to visualize the Danish corporate network, or the ones of political commissions etc.
Here, we make use of the following functions:
-
we use the
select()function to select specific columns, keeping all rows constant -
we use the
filter()function to select specific rows, keeping all columns constant
Both of these functions are used to make subsets, that are then assigned to new data objects. Below, we assign a subset of den which only looks at Corporations to the object den1.
# selecting the name and affiliation of a person
den %>%
select(name, affiliation)
## # A tibble: 56,849 × 2
## name affiliation
## <chr> <chr>
## 1 Aage Almtoft Middelfart Sparekasse
## 2 Aage B. Andersen Foreningen OEstifterne - Repraesentantskab (Medlemmer a…
## 3 Aage Christensen AARHUS SOEMANDSHJEM
## 4 Aage Dam Brancheforeningen automatik, tryk & transmission (besty…
## 5 Aage Dam Dansk Erhverv (bestyrelse)
## 6 Aage Frandsen Dommere valgt af Folketinget (Rigsretten)
## 7 Aage Juhl Joergensen Vestforsyning
## 8 Aage Krogsdam Danske Rejsejournalister (bestyrelse)
## 9 Aage Larsen Liberalt Oplysnings Forbund (LOF) (Landsstyrelsen)
## 10 Aage Lauridsen Halinspektoerforeningen (Bestyrelse)
## # ℹ 56,839 more rows
# filtering the data set to only show the sector of corporations
# and assigning it to the object den1
den1 <-
den %>%
filter(sector == "Corporations")
# look at the object den1
den1
## # A tibble: 7,989 × 17
## name affiliation role tags position_id id sector type description
## <chr> <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
## 1 Aage Alm… Middelfart… Memb… Corp… 1 95023 Corpo… <NA> Automatisk…
## 2 Aage Juh… Vestforsyn… Memb… Corp… 28 100249 Corpo… <NA> Automatisk…
## 3 Adam Chr… IF IT Serv… Exec… Corp… 131 92328 Corpo… <NA> Automatisk…
## 4 Adam Tro… Ejner Hess… Memb… Corp… 186 87369 Corpo… <NA> Automatisk…
## 5 Adam Tro… ISS FACILI… Chai… Corp… 187 92613 Corpo… <NA> Automatisk…
## 6 Adine Ch… HI3G Denma… Memb… Corp… 220 91906 Corpo… <NA> Automatisk…
## 7 Agnes Ma… Dalum Papir Chie… Corp… 302 85188 Corpo… <NA> Automatisk…
## 8 Agnete D… TRE-FOR (R… Memb… Corp… 321 110225 Corpo… Virk… <NA>
## 9 Agnete R… Broedrene … Chai… Corp… 353 84179 Corpo… <NA> Automatisk…
## 10 Agnete R… Novozymes Vice… Corp… 355 128276 Corpo… <NA> Automatisk…
## # ℹ 7,979 more rows
## # ℹ 8 more variables: created <dttm>, archived <dttm>, last_checked <dttm>,
## # cvr_person <dbl>, cvr_affiliation <dbl>, person_id <dbl>,
## # affiliation_id <dbl>, gender <chr>
All of these functions can be combined through the pipe operator which allows you to write two commands at once without having to assign it to an object in the meantime. How would you e.g. find the affiliation with the most members in the corporate sector?
# find a list of corporate affiliations and order them
den %>%
filter(sector == "Corporations") %>%
count(affiliation, sort = T)
## # A tibble: 1,195 × 2
## affiliation n
## <chr> <int>
## 1 TRE-FOR (Repraesentantskab) 119
## 2 Kromann Reumert 55
## 3 Bech-Bruun 54
## 4 Gorrissen Federspiel 40
## 5 Plesner 40
## 6 EnergiMidt 31
## 7 Lett Law Firm 27
## 8 Koebenhavns Lufthavns Vaekstkomité (Medlemmer) 24
## 9 Syd Energi (SE) 24
## 10 TDC (note) 24
## # ℹ 1,185 more rows
1.4 Creating graphs
Before we can make a network graph, we need to transform the data from a data frame into a data matrix.
There are two important kinds of data matrices of which networks are constructed:
-
A two-mode network is based on an
incidence matrixwhich is an matrix, where corresponds to one unit (such as an individual), and corresponds to another unit (such as organization). Each entry if there is an edge between node and node and otherwise. -
A one-mode network is based on an
adjacency matrixwhich is an matrix, where both row elements and column elements corresponds to nodes with the same unit. It is a symmetric matrix where each entry if there is an edge between node and node and otherwise.
In R, we create an incidence matrix by cross tabulating two columns of a data frame with the xtabs() function which stands for cross tabulation. To transform an incidence matrix into an adjacency matrix, we perform a simple matrix multiplication of the incidence matrix with the transposed version of itself.
## incidence matrix ##
# we use the argument sparse to save space on our memory because
# every time there is no edge, the compute will write a "." finstead of a 0
incidence <- xtabs(formula = ~ name + affiliation,
data = den1,
sparse = TRUE)
#show the first two rows and first two cols
incidence[1:2,1:2]
## 2 x 2 sparse Matrix of class "dgCMatrix"
## affiliation
## name & co 3C Groups
## Aage Almtoft . .
## Aage Juhl Joergensen . .
## Adjacency matrices ##
# individuals * individuals matrix
adj_i <- incidence %*% Matrix::t(incidence)
#show the first two rows and first two cols
adj_i[1:2,1:2]
## 2 x 2 sparse Matrix of class "dgCMatrix"
## Aage Almtoft Aage Juhl Joergensen
## Aage Almtoft 1 .
## Aage Juhl Joergensen . 1
# affiliation * affiliation matrix
adj_a <- Matrix::t(incidence) %*% incidence
#show the first two rows and first two cols
adj_a[1:2,1:2]
## 2 x 2 sparse Matrix of class "dgCMatrix"
## & co 3C Groups
## & co 4 .
## 3C Groups . 3
1.4.1 Loading graph objects
Graph objects can be loaded through a variety of ways. In this course, we concentrate on loading graph objects from incidence and adjacency matrices with the igraph package.
Since some people have several affiliations (see data set den), graph objects loaded from with the function graph_from_adjacency_matrix() contain loops and weights. In order to remove those loops and weights again, and to make the graph object more lean, we add the simplify() function.
# making a two-mode graph
gr <- graph_from_incidence_matrix(incidence, directed = FALSE)
## Warning: `graph_from_incidence_matrix()` was deprecated in igraph 1.6.0.
## ℹ Please use `graph_from_biadjacency_matrix()` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
gr
## IGRAPH 4bbb65e UN-B 8090 7989 --
## + attr: type (v/l), name (v/c)
## + edges from 4bbb65e (vertex names):
## [1] Lars Eivind Kreken --& co Mikael Ernst Joergensen --& co
## [3] Thomas Hoegeboel --& co Thomas Hoffmann --& co
## [5] Nicoline E. Hyldahl --3C Groups Niels Thorborg --3C Groups
## [7] Soeren Melanchton Pedersen--3C Groups Lone Lehmann Laurberg --3M
## [9] Maurizio Botta --3M Nicolas Nees Henriksson --3M
## [11] Niels Roeddik --3M Rasmus Gymoese Berthelsen --3M
## [13] Ulla Charlotte Ravn --3M Bo Boje Larsen --3xN
## [15] Jan Ammundsen --3xN Jeanette Hansen --3xN
## + ... omitted several edges
# one-mode graph for individuals
gr1 <- graph_from_adjacency_matrix(adj_i, mode = "undirected") %>% simplify(remove.multiple = TRUE, remove.loops = TRUE)
# one-mode graph for affiliations
gr2 <- graph_from_adjacency_matrix(adj_a, mode = "undirected") %>% simplify(remove.multiple = TRUE, remove.loops = TRUE)
Having loaded the graph objects, we are able to visualize them using the ggraph package.
1.5 Visualizing network graphs

The syntax for plotting network graphs in the ggraph package is very close to the ggplot package. We start out with the ggraph() function, where we specify the graph object. This creates a plain board which can be filled accordingly with other functions. Here is an example code for visualizing two-mode networks.
# Visualize the two mode graph
gr %>%
# there are different layout types which changes the position of the nodes. For now, we only use this one called "kk" or "fr"
ggraph(layout = "kk") +
# draws edges
geom_edge_link0(color = "black", edge_width = 0.1) +
# draws nodes
geom_node_point(aes(color = type), size=0.2, alpha = 0.4) +
# # draws labels for type==TRUE and repels the overlapping ones.
# geom_node_text(aes(filter=type==TRUE, label =name), size=0.8, repel = TRUE) +
# overrides the existing legend
scale_color_manual(values=c("sienna1", "steelblue2"), labels=c("individuals", "companies")) +
theme_graph()
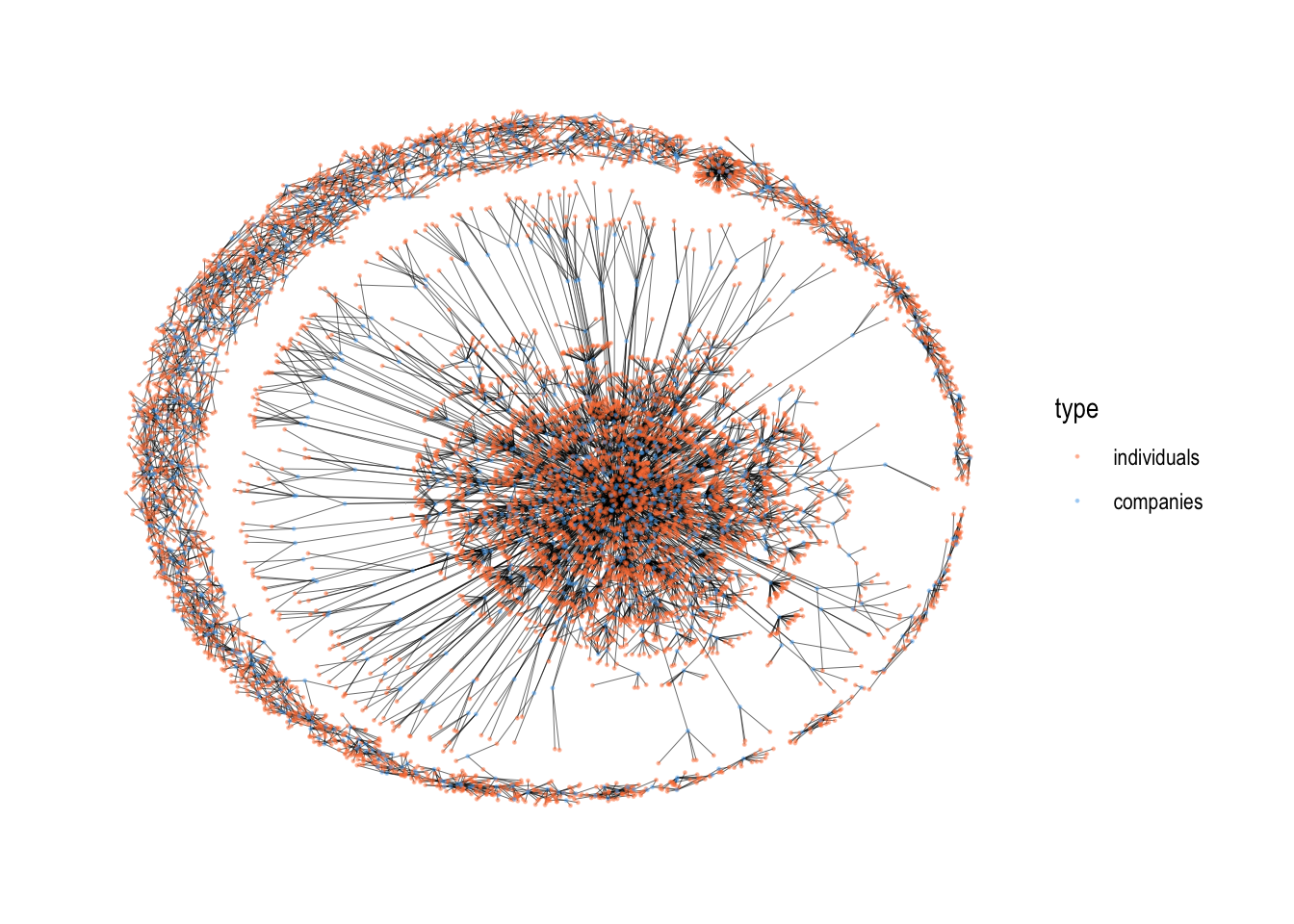
Despite the fact that we reduced the node size drastically, it is still difficult to view this network because of the limited page size. After all, it is a network with 8,090 nodes and 7,989 edges.
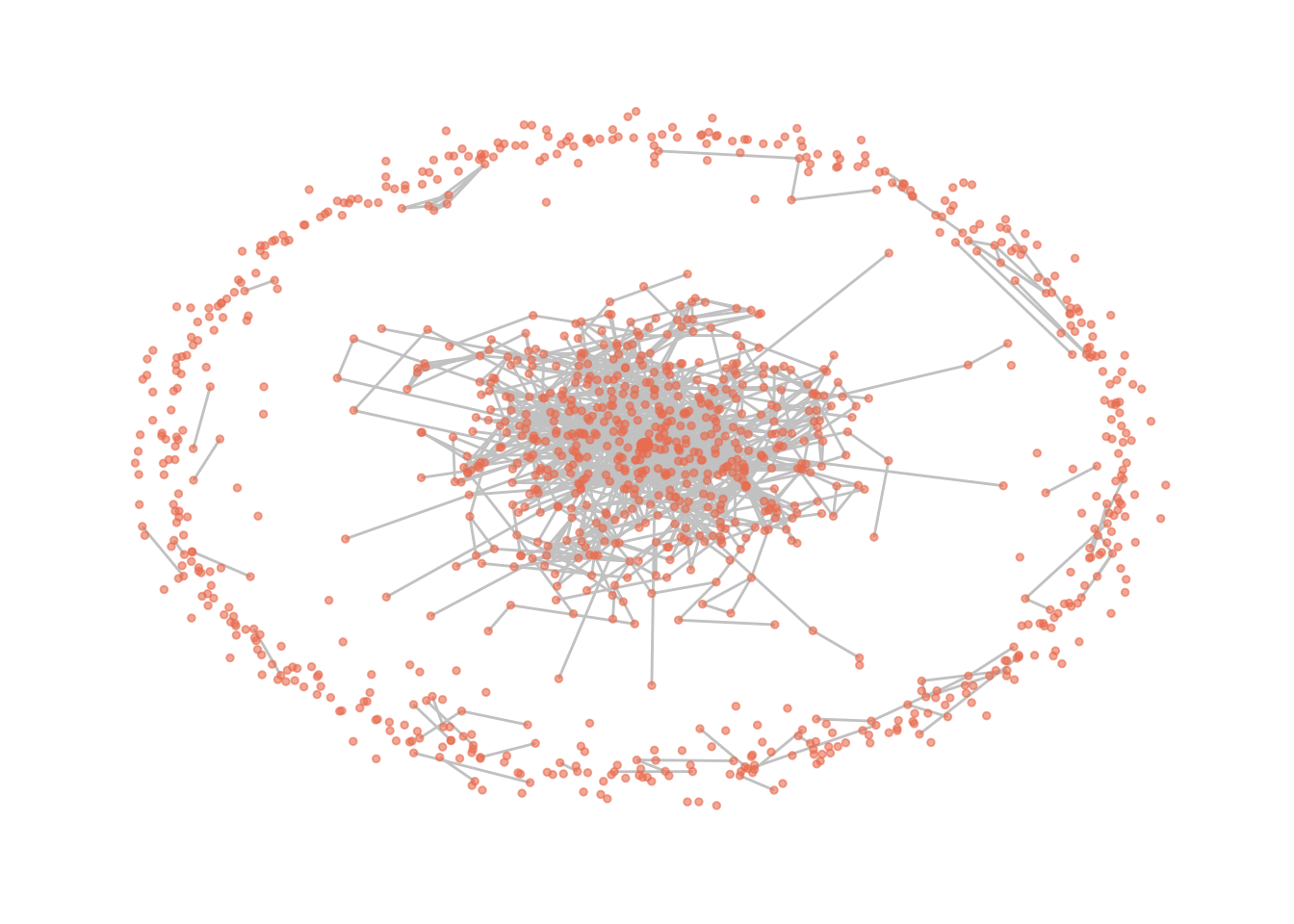
Let us also visualize the one-mode network of companies.
# Visualize the two mode graph
gr2 %>%
# # there are different layout types which changes the position of the nodes. For now, we only use this one called "kk" or "fr
ggraph(layout = "kk") +
# draws edges
geom_edge_link0(color = "gray80") +
# draws nodes
geom_node_point(size=1, alpha = 0.6, color = "salmon2") +
theme_graph()
R-scripts
To practice a little more, you can download an empty exercise sheet. You can also find a filled out cheat sheet below.